Once considered a type of gout, rheumatoid arthritis (RA) is now known as a prototypical autoimmune disease with undefined pathogenic diversity. New research led by physician-scientists at the Brigham and Women’s Hospital Division of Rheumatology, Inflammation, and Immunity is shifting the perspective on RA again, providing novel insights into the disease’s pathology and helping to advance targeted drug development and predict treatment response.
The research, conducted under the National Institute of Health’s Accelerating Medicines Partnership Program: Rheumatoid Arthritis and Systemic Lupus Erythematosus Network, involved a consortium of members from leading academic medical centers around the world.
Michael B. Brenner, MD, director of cell and molecular immunology in the Division, led the working group that developed the concept for the consortium and the group’s various sections. Meanwhile, Brigham investigator Soumya Raychaudhuri, MD, PhD, led the group’s systems biology effort and analyzed all of the research data generated. The consortium’s research was recently published in Nature.
The two colleagues share an interest in developing treatments, therapies, and cures for RA.
“Genes play specific roles in specific cell states,” Dr. Raychaudhuri says. “If we know the right genes and cell states to target within inflammatory tissue, researchers can prioritize them for functional investigation and develop more effective therapies.”
Single-Cell RNA Sequencing Builds on Earlier Cytometry and Histology Work
The Nature study, which used RNA sequencing to provide single-cell disease deconstruction of biopsied synovial tissue from RA patients, provides an amplified magnitude of detail over previous single-cell analyses that used flow cytometry and histology to identify surface markers on RA tissue cells.
The genetic component, Dr. Brenner notes, is important because genes expressed in the cell act as a surrogate for what the cell is doing. “We can redefine the cells present by type and state, identify their behaviors, and understand the functions they carry out,” he adds.
Through a single-cell atlas of RA synovial tissue that includes more than 314,000 cells, the research consortium stratified tissues into six groups, called cell-type abundance phenotypes (CTAPs), each characterized by selectively enriched cell states.
“These CTAPs demonstrate the diversity of synovial inflammation in rheumatoid arthritis, ranging from samples enriched for T and B cells to those largely lacking lymphocytes,” the study authors state. “Disease-relevant cell states, cytokines, risk genes, histology, and serology metrics are associated with particular CTAPs.”
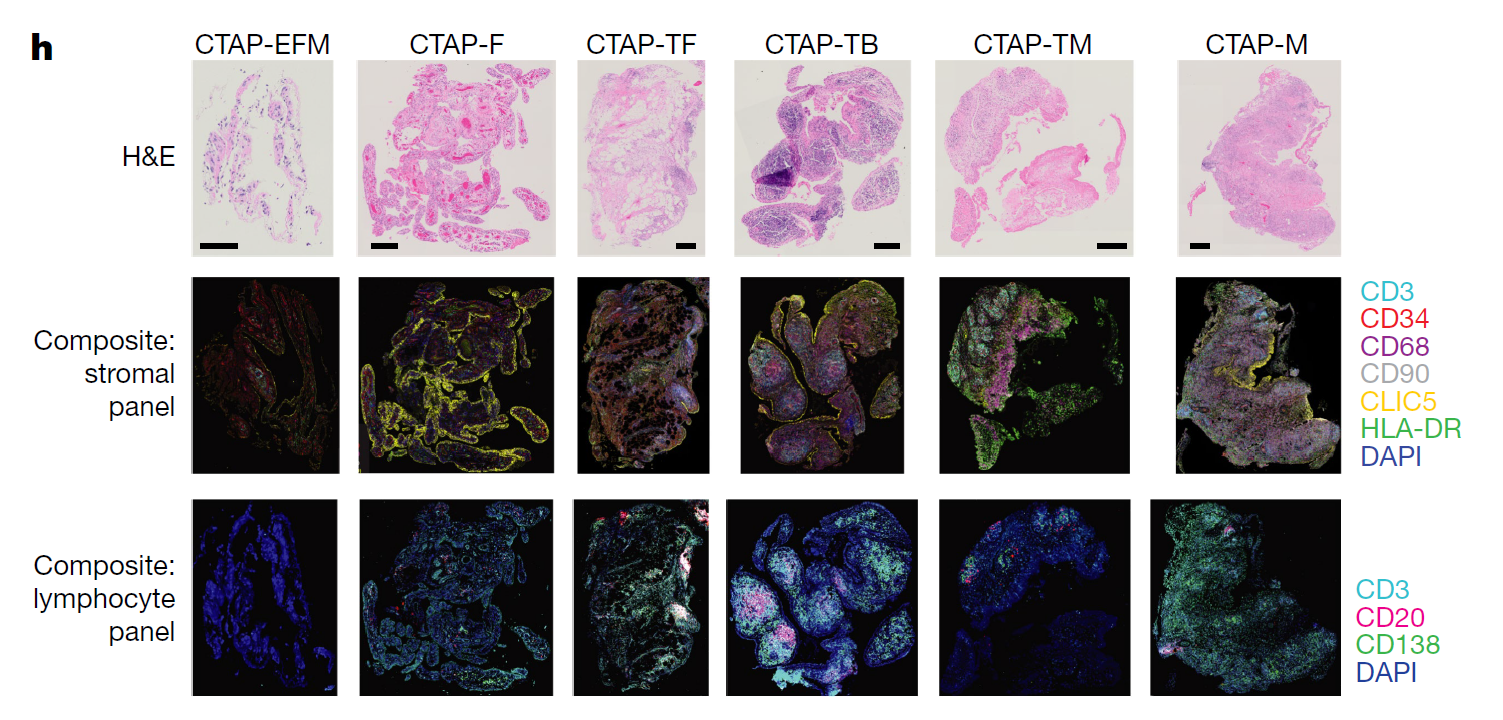
Figure 1h: “Representative synovial tissue fragments from each of the CTAPs. Top row, haematoxylin and eosin (H&E) staining. Middle row, immunofluorescence microscopy for CD3, CD34, CD68, CD90, CLIC5, and HLA-DR. Bottom row, immunofluorescence microscopy for CD3, CD20, and CD138.” Figure and caption published in Nature.
Stratifying RA Patients for Targeted Treatment Development
According to Dr. Brenner, the discovery of the new cell types and cell states was one of the most exciting results of the study. It came about, he says, through the compilation and integration of vast datasets with hundreds of thousands of data points that allowed the consortium’s researchers to stratify patients based on the pathology of their synovial tissue.
“RA was previously considered to be ‘one’ disease,” Dr. Brenner says. “But this study showed six or more different pictures of the cellular composition of tissue pathology. In some patient samples, we predominantly saw lymphocytes like T cells and B cells. In other patients, we saw mainly myeloid macrophages and monocytes. And in others, we predominantly saw fibroblasts and other stromal populations.
“This ‘one’ disease is now proven to have at least six different subtypes, allowing us to redefine the pathology of the disease.”
The study included data from both patients naïve to treatment and those who failed treatment. Many treatment non-responders were found to have a proliferation of fibroblasts in their synovial tissue, providing new insights into a disease largely thought to be driven by T cells, B cells, and myeloid cells.
“We previously thought that as RA symptoms improve, we would see fewer T cells and B cells relative to tissue cells,” Dr. Raychaudhuri says. “But in fact, there is a subset of patients with active disease who have more fibroblasts and fewer T cells and B cells and are less likely to respond to conventional treatment. These patients represent an important subgroup, and it may be clinically valuable to identify them in advance and to develop specialized therapies for them.”
Next Step: Studying Cell-cell Interactions
Drs. Brenner and Raychaudhuri plan to continue their consortium work in RA cell states through a new partnership focused on cell-cell interactions. Using spatial cell analysis, they seek to discover which cells send signals to other cells to create the network of immune system reactions resulting in RA pathology.
“Instead of looking at thousands of genes within a cell, we will look at thousands of genes within the actual tissue space,” Dr. Raychaudhuri says. “When we see specific cell states next to each other and interacting with each other, we will be able to determine whether that interaction is driving the pathology.”
The consortium’s efforts, including parallel work in lupus nephritis on kidney single-cell deconstruction, is a testament to the power of team science and the Brigham’s deep bench in basic science, computational biology, translational investigation, and clinical expertise.
“The concept of a consortium like this isn’t typical in the inflammatory disease field, let alone one that was funded with $42 million over eight years,” Dr. Brenner says. “We were honored to play an integral role in a worldwide network of scientific investigators dedicated to pushing the field of rheumatology forward in meaningful ways.”