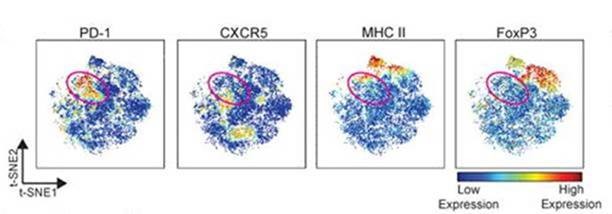
A myriad of “single cell” technologies now offer the potential to redefine our understanding of human immunology. Traditional approaches to the examination of individual cells have included flow cytometry, which has enabled pathologists and immunologists to assess up to a maximum of 12 protein markers on a given cell. Over the decades these technologies have allowed scientists to characterize individual cell populations of CD4+ T cells, B cells, and other immune populations, which are essential in the pathophysiology of rheumatoid arthritis and other rheumatic diseases. But now new single cell technologies, such as single cell RNA-seq, allow investigators to query the expression of thousands of genes simultaneously.
These newer technologies possess extraordinary potential to unlock the secrets of rheumatic diseases. At Brigham and Women’s Hospital (BWH), Michael B. Brenner, MD, chief of the Division of Rheumatology, Immunology and Allergy and Soumya Raychaudhuri, MD, PhD, are now using these technologies to assay not only individual immune cells from blood, but also immune and stromal cells in the synovial tissue of rheumatoid arthritis patients.
As part of the NIH and industry-funded Accelerating Medicines Partnerships (AMP) program, BWH investigators are leading a consortium of investigators throughout the United States and the United Kingdom to secure specimens of blood and synovial tissues from hundreds of patients. Working with the AMP, Drs. Brenner and Raychaudhuri are applying single cell technologies to these samples to investigate how disease progresses, which cell populations correspond to the presence of rheumatoid arthritis, which populations expand with disease progression, and which ones portend the best and worst clinical response to therapies.
The data sets generated from this study are enormously complex, and require novel algorithms that can account for the thousands of genes queried in thousands of cells in an individual experiment. Dr. Brenner and his team lead the tissue processing and immunological investigation of these samples. Dr. Raychaudhuri and his team lead the development of computational strategies to help define the populations, and their key genetic alterations. This remarkable team is melding experimental and computational science in a uniquely powerful way for the study of rheumatic diseases.
Along with inflammatory arthritis, overproduction of autoantibodies is a defining feature of rheumatoid arthritis (RA), particularly autoantibodies against citrullinated proteins (ACPAs). The clinical significance of these ACPAs, other than for diagnosing RA, has been unclear. Previous studies have focused on association studies between a few ACPAs and one or two specific RA phenotypes – for example, an association between anti–citrullinated histone H2B antibodies and coronary artery calcium scores in patients with RA. But such an approach cannot identify potential associations with a wider array of untested phenotypes.
In a recently published article (Arthritis Rheumatology, 2017 Apr; 69(4): 742–749), Dr. Katherine Liao and colleagues applied the Phenome Wide Association Study (PheWAS) approach to screen for associations between ACPAs and potential subphenotypes of RA.
A PheWAS can be viewed as a Genome-Wide Association Study (GWAS) turned on its head. In a standard GWAS, an investigator tests for association between approximately one million genetic variants and one phenotype outcome, such as RA, yes or no. Using a standard PheWAS, it is possible to test for associations between genetic variants and a broad range of phenotypes.
The PheWAS generates a broad range of testable hypotheses and can be applied in large biobanks linked with electronic medical record (EMR) data. A published mapping converts groups of ICD9 codes into phenotypes. For example, RA would be defined as ICD9 714.x – ‘Rheumatoid arthritis and other inflammatory polyarthropathies’. It was believed that the PheWAS could be an effective approach to examine whether the ACPAs differentiate RA into clinically relevant subgroups. Because the PheWAS was designed for genetic data, an interdisciplinary team of biostatisticians and bioinformaticians developed new methods that allowed screening for associations between groups of biomarkers – namely, ACPAs – and multiple phenotypes.
This study was conducted in an RA cohort created using EMR data and linked to specimen samples that were measured for 36 autoantibodies with a published multi-bead assay. The autoantibodies were then grouped by their target protein for instance, the 10 antibodies against fibrinogen comprised one group. ICD9 codes for all RA subjects were then extracted and grouped into phenotypes using published PheWAS grouping methods.
The analysis included 1,006 RA patients, 10 groups of autoantibodies, and 625 phenotypes, and identified an association between anti-citrullinated fibrinogen with ischemic heart disease, which confirmed an earlier study that observed an association between anti-cit-fibrinogen and atherosclerotic plaque burden (Figure). Additionally, a strong association between autoantibodies against fibrinogen with inflammatory lung conditions was identified. Some of the strong associations found in this study are under investigation in prospective studies as potential biomarkers of risk for outcomes in coronary artery disease and interstitial lung disease among RA patients.
Big data will drive precision medicine in rheumatology. At Brigham and Women’s Hospital (BWH), the combination of massive databases with genomic and clinical data will open new capabilities for the diagnosis, care, and even prevention of rheumatic disease.
BWH’s investment in large-scale data collection is bearing fruit. The Partners HealthCare Biobank repository of DNA, plasma, and serum samples now has more than 78,000 participants, with 20,000 genotyped samples, as well as survey data with family history, lifestyle and environmental information. In 2015, BWH and Partners joined the NIH-funded Electronic Medical Records and Genomics (eMERGE) Network, one of 11 sites in the United States that combines biorepositories such as the Biobank with data gathered from electronic medical records (EMRs).
Together, the Biobank and eMerge create robust tools for linking genomic data to phenotype, providing greater understanding of how genes and environmental factors influence health and disease.